Researchers in New York say their new ‘programmable antibiotic’ treatment that fights dangerous bacterial infections without harming other more benign or even helpful microbes shows promise.
The technique could also some day prove to be a useful tool in the fight against the growing global health threat of antimicrobial resistance
In a study published in the journal Nature Biotechnology, the scientists outlined a new treatment technique that will allow antibiotics to be customized and specially programmed to go after bad germs without harming the good bacteria that help our bodies function properly.
“In experiments, we succeeded in instructing a bacterial enzyme, known as Cas9, to target a particular DNA sequence and cut it up,” said lead researcher Luciano Marraffini, head of Rockefeller University’s Laboratory of Bacteriology in a press release.
“This selective approach leaves the healthy microbial community intact, and our experiments suggest that by doing so you can keep resistance in check and so prevent certain types of secondary infections, eliminating two serious hazards associated with treatment by classical antibiotics,” said Marrafini.
Traditional antibiotics go after harmful bacteria with such force that they often take out many of the “good microbes” while attacking the “bad microbes”.
Bacteria can mutate in a way that lets them develop an immunity to standard antibiotic treatments. As a result, medical science has been developing stronger and stronger medications to fight these mutating microbes.
To seek out and destroy certain DNA sequences in harmful bacteria, but leave others alone, the researchers adopted an immunity system, called CRISPR – Clustered Regularly Interspace Short Palindromic Repeats – that bacteria use to protect themselves.
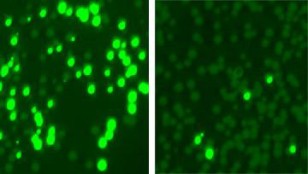
Rockefeller University researchers colonized mouse skin with a mix of bacterial cells, some resistant to the antibiotic kanamycin. They made the resistant cells glow (left) and treated the mix with an enzyme that targeted and killed off most resistant cells (right). (Marraffini Lab and Fischetti Lab/Nature Biotechnology)
Contained within these CRISPR systems are some unique genetic sequences called spacers that match up with the sequences found in intruding viruses.
Among the enzymes that are linked with the system is the CRISPR associated protein 9 or Cas9 enzyme.
The New York research team was able to send the Cas9 enzyme to locate and destroy a target within a particular strain of a harmful bacteria by engineering the CRISPR spacer sequences so that it matched the “bad microbe’s” genes.
These engineered sequences, along with Cas9 enzyme, were inserted into a cell of a “bad microbe”. Once inside, the combination causes the cell to turn on its own immunity system. Depending on where its target was located within the cell, the Cas9 could wipe out the target gene or destroy the cell itself.
The researchers also found that in some instances, this type of treatment could prevent a harmful microbe from developing any resistance at all.
“We previously showed that if Cas9 is programmed with a target from a bacterial genome, it will kill the bacteria. Building on that work, we selected guide sequences that enabled us to selectively kill a particular strain of microbe from within a mixed population,” says first author David Bikard, a former Rockefeller postdoctoral research who is now at the Pasteur Institute in Paris.
To help reach their findings, the researchers conducted three sets of experiments.
The first involved a common skin and respiratory microbe that can resist the antibiotic kanamycin. In this situation, the Cas9 targeted and killed the antibiotic resistant portion of the bacteria, leaving the rest to be destroyed by the kanamycin antibiotic.
In the second set of experiments the researchers sent the Cas9 after some tetracycline-resistant elements in a strain of potentially deadly bacteria known as Methicillin-resistant Staphylococcus aureus or MRSA. The Cas9 treatment in this case not only allowed the bacteria to again be vulnerable to tetracycline again, but it also set up other staph cells to act as a form of immunization that prevented the bacteria from adopting elements that made it resistant to antibiotics.
In the third and final set of tests the researchers used the Cas9 to selectively destroy antibiotic resistant staph infections from the shaved backs of mice.
The researchers said that although their work produced promising results, the delivery method used to carry out this new treatment needs further improvement.



















Comments are closed.